A common format for federated learning: CAIA’s approach to data standardization
Whenever a patient visits their healthcare provider, they begin a complex “diagnostic odyssey.” Hospitals document a wide range of information including scans, blood work, doctors’ notes, and treatment options. From a patient’s perspective, it may seem like all records look the same across major hospitals. The reality, however, is that every institution has a different way of recording this information, often referred to as clinical data, creating a significant challenge for collaborative cancer research.
In its first year, the CAIA team set out to solve this challenge: standardizing data structure and formats at participating cancer centers to better unlock research insights. CAIA’s federated learning approach ensures that researchers only have access to de-identified clinical data, a crucial regulatory requirement — we explain how we adhere to this requirement below. However, the de-identified data structure and format need to be uniform and accessible for researchers to work seamlessly.
In this blog post, we take a look at the data standardization process, and how CAIA adheres to privacy, regulatory, ethical and legal guidelines. This process will help participating cancer centers develop advanced, equitable, and diverse AI models that have the potential to uncover new patterns and shed light on new therapies.
What is data standardization?
We know that hospitals gather a substantial amount of data on every patient’s health. Now, imagine combining a single patient’s data, with millions of other patient journeys across the country, to create powerful AI models to find new cures.
The key to unlocking this potential is data standardization. This work ensures that the vast amount of information collected during a patient's care can be interpreted, and reliably analyzed across different institutions. In other words, the goal of data standardization is to create a common format that is accessible and useful to researchers for collaborative, multi-institutional cancer discovery.
De-identifying patient data to protect privacy
For CAIA’s participating cancer centers, protecting patient privacy is a paramount concern. Complying with human subjects research and privacy regulations is crucial. A key step in the compliance process is de-identification of patient records. De-identifying patient data is a complex process because the data must not contain identifiable information while still retaining its usefulness for research.
Here are couple of examples of how patient privacy is protected:
Removing information which by itself, or in combination with other information, may reveal an individual patient’s identity.
Shifting dates to preserve the ability to study time-dependent events—like progression-free survival (time from treatment to disease progression). Researchers don’t simply remove all dates. Instead, they shift every single date for an individual patient by a random, consistent number of days. This preserves the relative sequence of events while rendering the patient virtually unidentifiable.
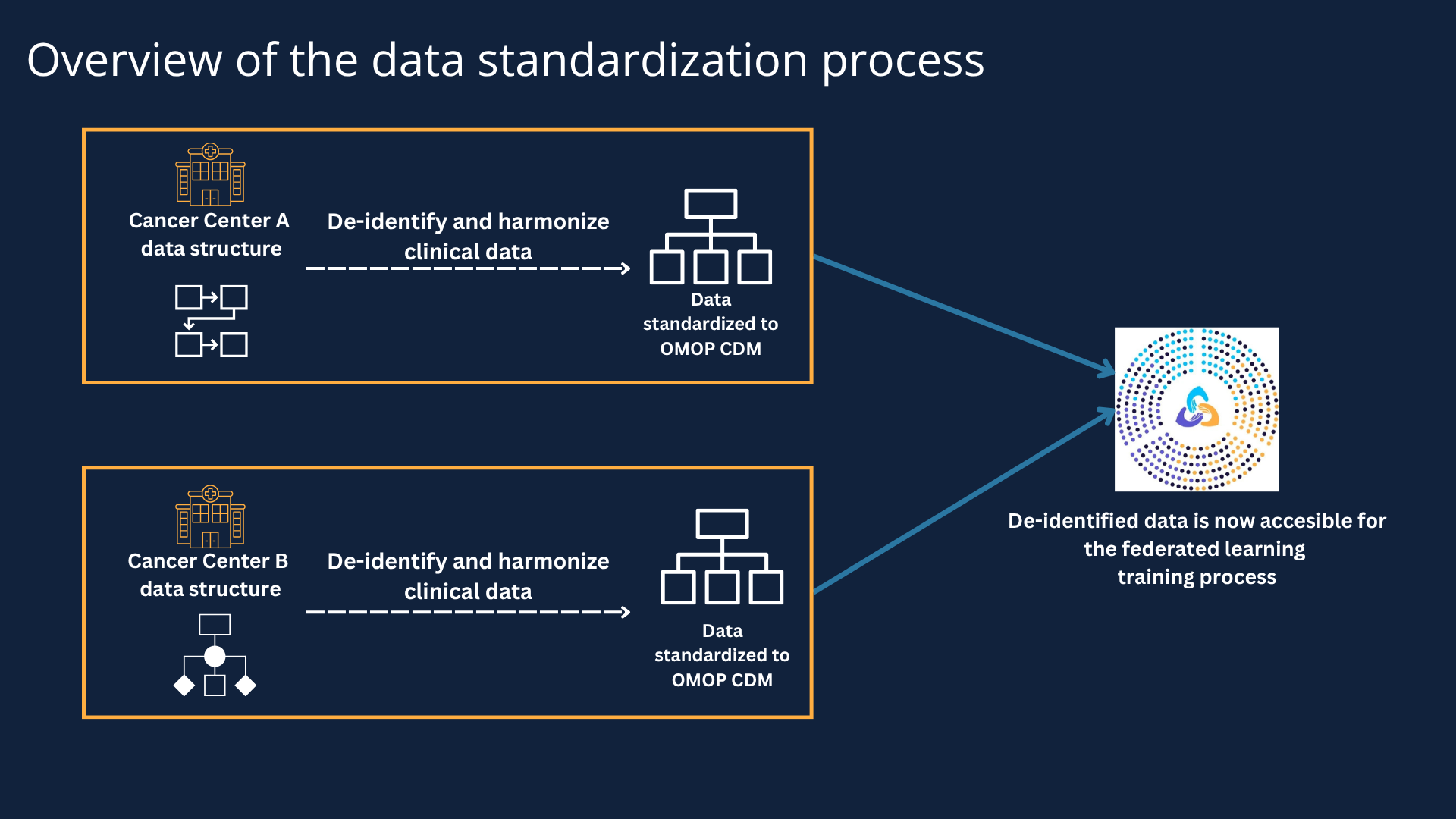
Overview of the data standardization process
Adopting a common vocabulary for standardization
John Methot, Vice President of Computational & AI Services at Dana-Farber Cancer Institute (one of CAIA’s founding cancer centers), has been central to CAIA’s data standardization work. As he explains, when a patient visits a cancer center, their experience generates two main types of data:
Structured Data: Clean, organized entries like the dose of a drug or the results of a lab test.
Unstructured Data: Free-text documents like pathology reports or radiology reports
Let’s now take a look at how CAIA standardized structured data across its participating cancer centers.
While each cancer center has their own data systems, CAIA still needed a way to map local coding systems and field names into a single, standardized format. A local coding system refers to a unique set of codes that hospitals use to track different procedures, lab results, and medications. If a researcher needs to run a multi-institutional study, they would need to know the local codes — and what they mean — for each participating center. So, these local codes have to be mapped to a universal, standardized system or a Common Data Model (CDM).
A Common Data Model (CDM) provides a framework for how different types of patient information and codes must be organized and structured. CAIA has adopted OMOP (Observational Medical Outcomes Partnership) as its core CDM. OMOP is an open data community standard that can translate local hospital codes into a universal standardized system, making the data understandable across all sites that share this same data model.
A good way to visualize a CDM is to imagine a large spreadsheet with many columns of information organized in a common language using standardized terms like “Condition Occurrence” or “Drug Exposure” or “Procedure Occurrence." Data from each cancer center is then slotted into each of these columns. For instance:
Diagnoses go into a "Condition Occurrence" column.
Prescribed medications go into a "Drug Exposure" column.
Procedures (like a biopsy or a surgery) go into a "Procedure Occurrence" column.
In short, adopting OMOP as CAIA’s Common Data Model will enable AI models to interpret the collective experience of millions of patients across institutions and provide researchers with standardized data that can be applied in a wide variety of clinical environments.
Beyond structured data
As we pointed out earlier, structured data is only one type of data collected during a patient’s care journey. However, this data provides essential context by linking diagnostic, treatment, and outcome information to other data types allowing for a complete understanding of a cancer patient’s condition and care trajectory. CAIA’s work with structured data will provide the infrastructure and “backbone” for connecting other kinds of data including medical imaging, genomic testing, and other aspects of routine care.
Training AI on multimodal data is the future. CAIA will continue to standardize more types of data to enable this and build the next generation of highly accurate AI models for precision cancer medicine
If you’d like to learn more about CAIA, subscribe to our newsletter for updates and follow us on LinkedIn and X.